Individual genomes harbor structural variations, such as insertions and deletions ranging from several hundred to millions of base pairs. Although most identified variations have biological impact and are of evolutionary significance, technical limitations have not allowed the study of SVs on the whole-genome or population level until very recently.
The fascination for plant genomics and the ability to adopt early to novel promising genomic tools allows us to be at the forefront of structural genomics. We are currently utilizing a BioNano Genomics Irys platform, Oxford Nanopore Tech MinION (both in-house) and PacBio SMRT sequencing to unravel the variable genome structure on the species level in Arabidopsis thaliana. The ultimate goal of the project, is to understand the changes in genome structure, and how geographic and ecological origin have shaped the chromosomal landscape.
The BioNano Genomics Irys platform allows us to create and compare physical genome maps for a large set of ecologically diverse Arabidopsis thaliana wild accessions, but also structured populations and mutants. Genome maps for Col-0 CS70000, on which the TAIR10 reference genome sequence is based, identified 29 regions of mis-assembly in highly repetitive genome regions in the chromosome arms. These regions are between 3 to 59 kb in size. Comparisons between wild accessions identifies conserved sites and types of SVs, mainly around the centromere, and evolutionary hotspots like NLR-type disease resistance gene clusters. We are also exploring the Irys platform for various options to label specific genomic sites as well as DNA modifications.
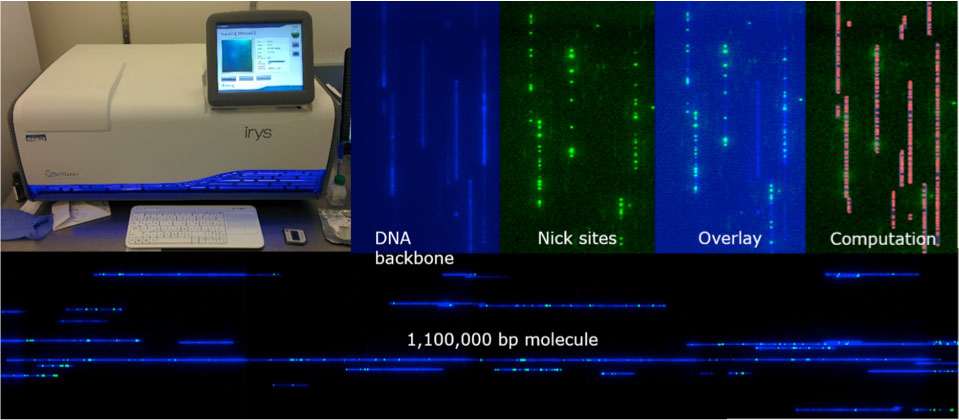
The combination of physical Irys maps and PacBio SMRT sequencing further allowed us to hybrid assemble all five chromosomes of A. thalianaLandsberg erecta (Ler-0) from Telomere to Telomere including rRNA and Telomere caps at chromosomes 2 and 4. Irys maps were used to order and close PacBio scaffolds around the centromeres. Gained sequence information provides unprecedented insights of the centromeric structure, size and repeat vs. non-repeat content.

The Oxford Nanopore MinION is currently used to sequence ultra-long DNA fragments that facilitate a relatively cheap scaffold for mapping the BioNano Genomics Irys physical maps. High error rate and low throughput are not inhibiting this workflow.