Science Snippets
Mobile Methylomes in Plant

Charting plant hormone signaling pathway networks using ChIP-Seq
Systems Biology of Hormone Regulated Seedling Growth
Telomere-to-telomere chromosome assemblies and identification of structural variations in Arabidopsis thaliana ecotypes

Center of Excellence in Stem Cell Genomics
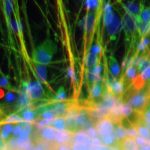
Epigenomic diversities of brain cell types
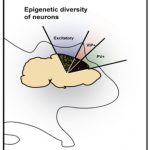
Roadmap Epigenome Project
ENCODE – Epigenetics During Developmental Trajectory

Evaluating Pluripotent Human Cells